Basic-Use
Basic-Use.RmdSimple Depiction
A simple wrapper around cdk’s excellent depict module.
library(depict)
library(magrittr)
# load a smiles string
pen <- parse_smiles("CC1(C(N2C(S1)C(C2=O)NC(=O)CC3=CC=CC=C3)C(=O)[O-])C penicillin")
# `depiction` is the base class; styles are applied to it
# `depict` applies the styles to the molecule
depiction() %>%
depict(pen) %>%
get_image() %>%
grid::grid.raster()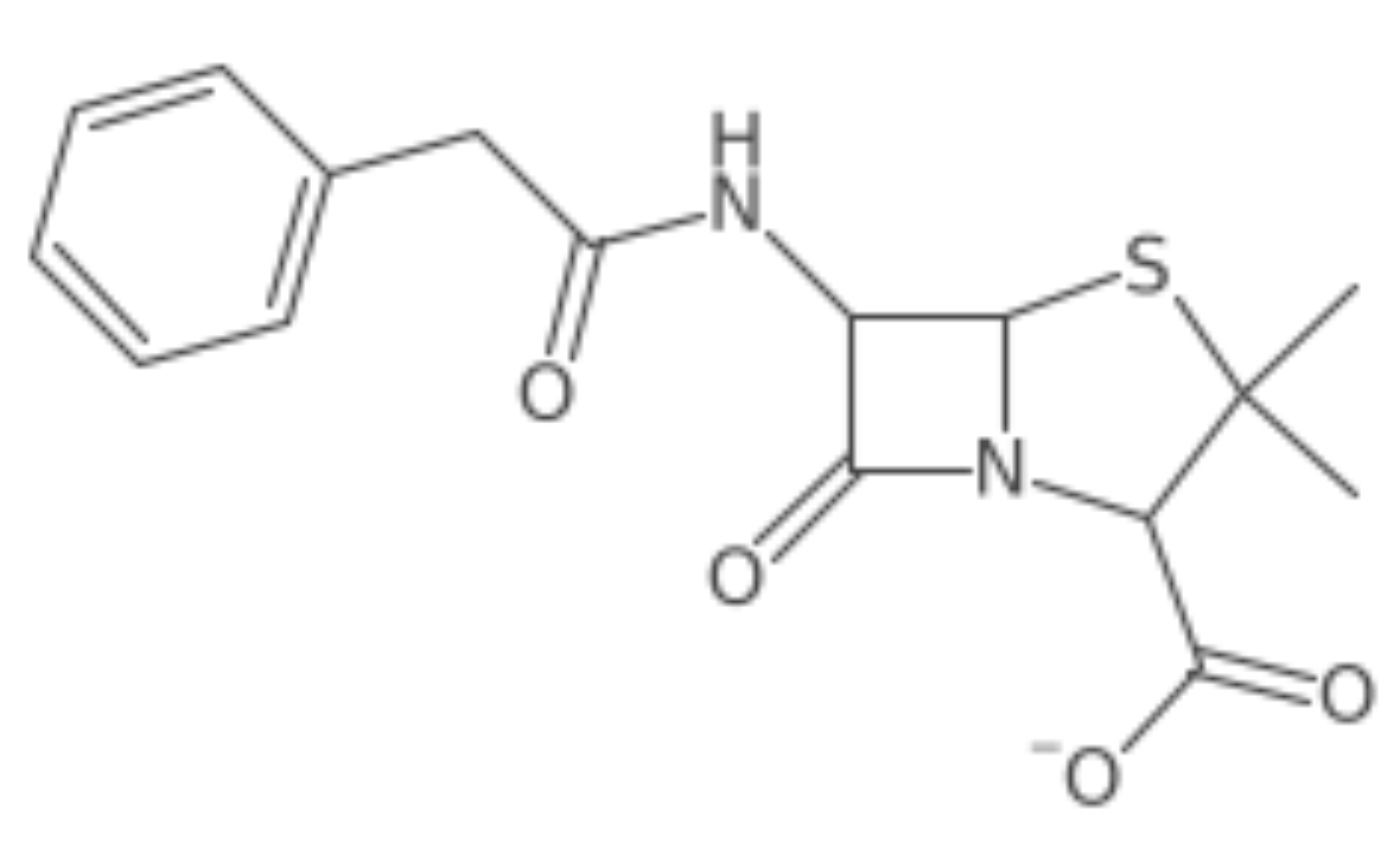
Multiple Smiles
# load a few smiles as AtomContainers
atmcontainers <- purrr::map(
c("CCCC", "CCCCCC", "CCCC1CCC1CC2CCCC2"),
parse_smiles)
# convert them into a java iterable
many_containers <- atomcontainer_list_to_jarray(atmcontainers)
depiction() %>%
set_zoom(10) %>%
depict(many_containers) %>%
get_image() %>%
grid::grid.raster()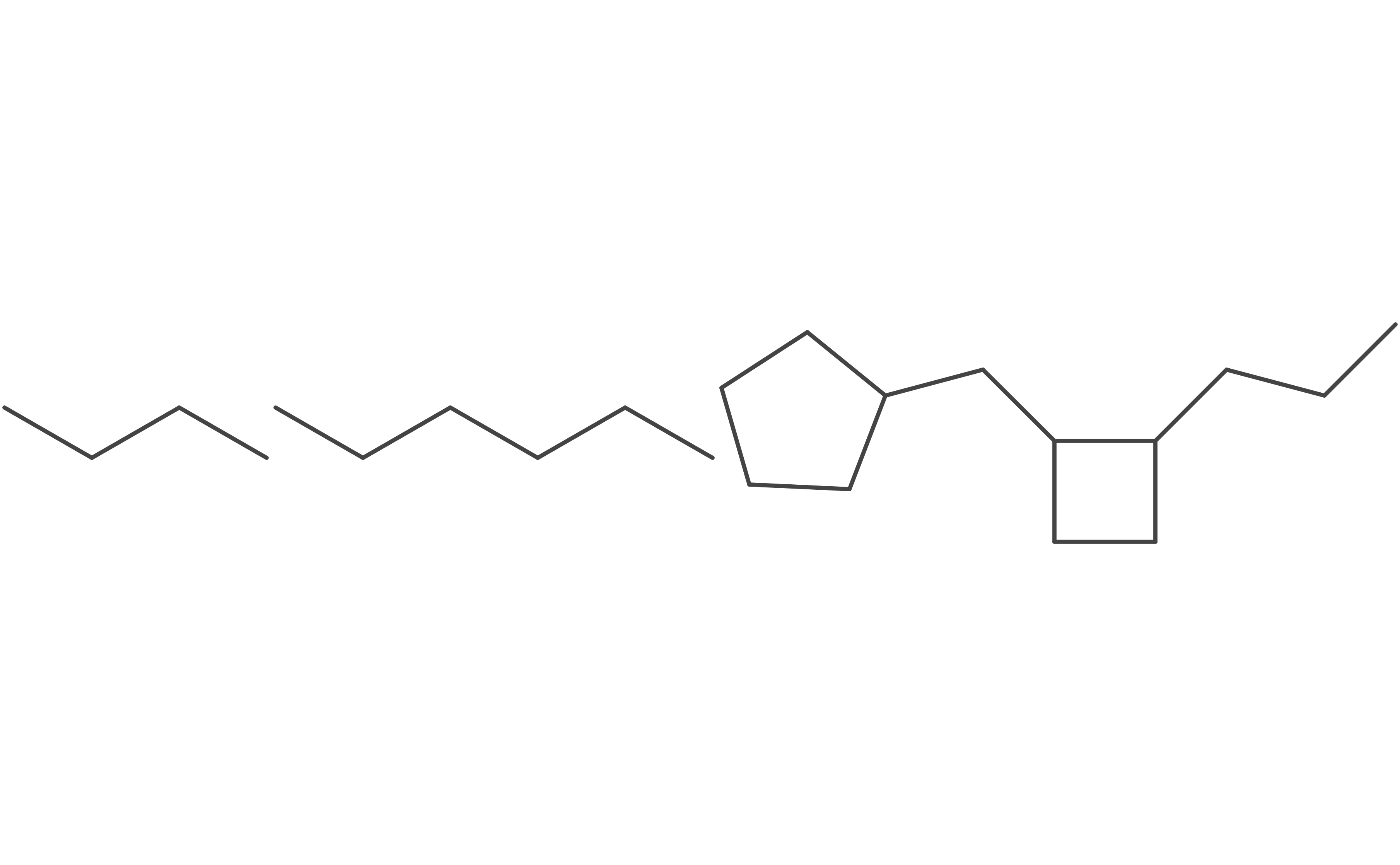
Use Colors, SMARTS, Highlighting
# you must supply java colors
color <- J("java.awt.Color")
# load in penicillin
pen <- parse_smiles("CC1(C(N2C(S1)C(C2=O)NC(=O)CC3=CC=CC=C3)C(=O)[O-])C penicillin")
cav <- parse_smiles("CN1C=NC2=C1C(=O)N(C(=O)N2C)C")
# define the regions to highlight
# either all atoms/bonds
# or a SMARTS-defined subregion
atms <- pen$atoms()
bnds <- pen$bonds()
lactam <- match_smarts("C1(=O)NCC1", pen)
# use piping to change the behavior as desired
depiction() %>%
highlight_atoms(atms, color$LIGHT_GRAY) %>%
highlight_atoms(bnds, color$LIGHT_GRAY) %>%
highlight_atoms(lactam, color$RED) %>%
set_size(400, 400) %>%
set_zoom(3) %>%
outerglow() %>%
add_title() %>%
depict(pen) %>%
get_image() %>%
grid::grid.raster()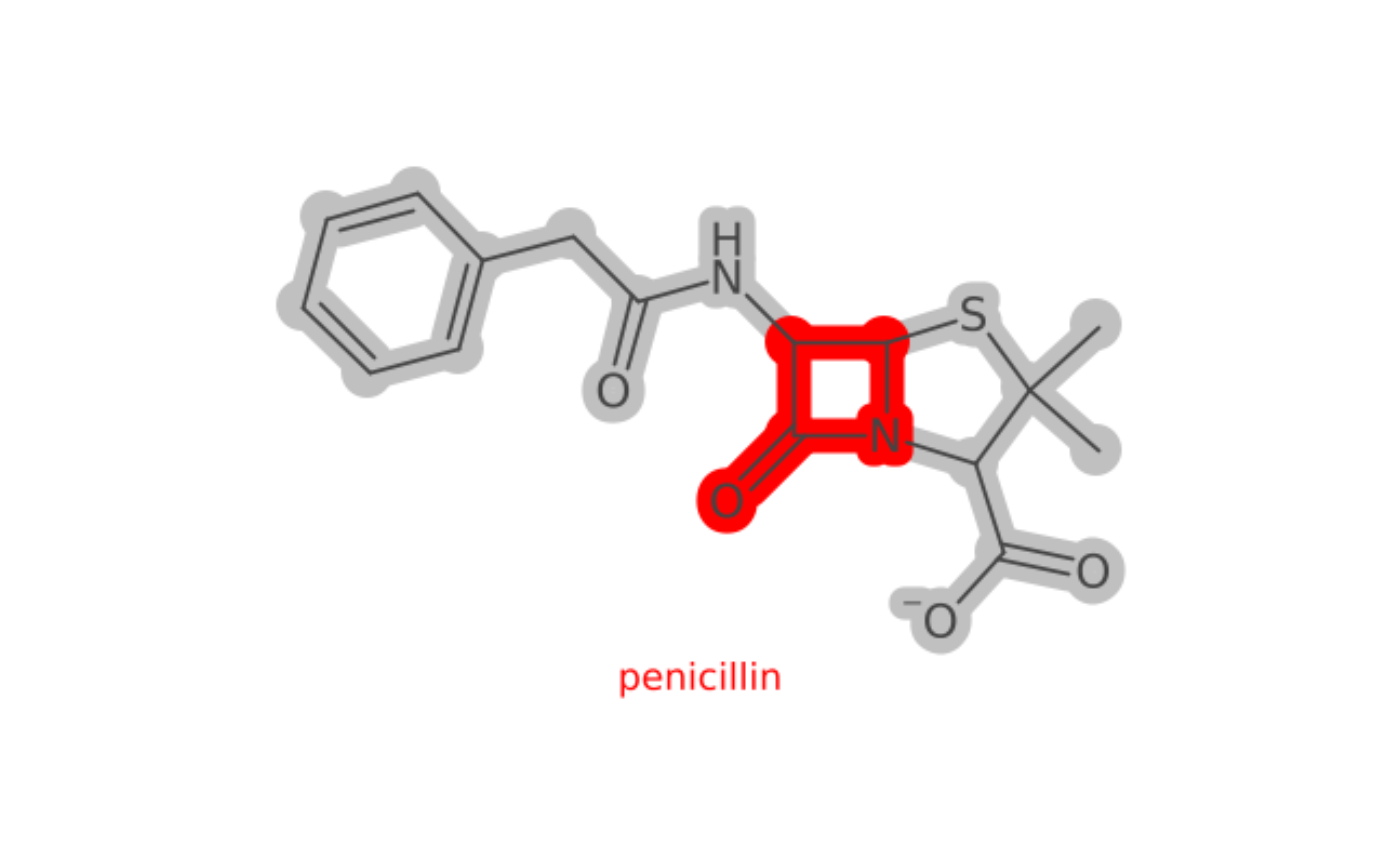
SMARTS for highlighting mutiple containers
small_highlight <- match_smarts("C1CCC1", many_containers)
depiction() %>%
highlight_atoms(small_highlight, color$RED) %>%
set_size(400, 400) %>%
set_zoom(3) %>%
outerglow() %>%
add_title() %>%
depict(many_containers) %>%
get_image() %>%
grid::grid.raster()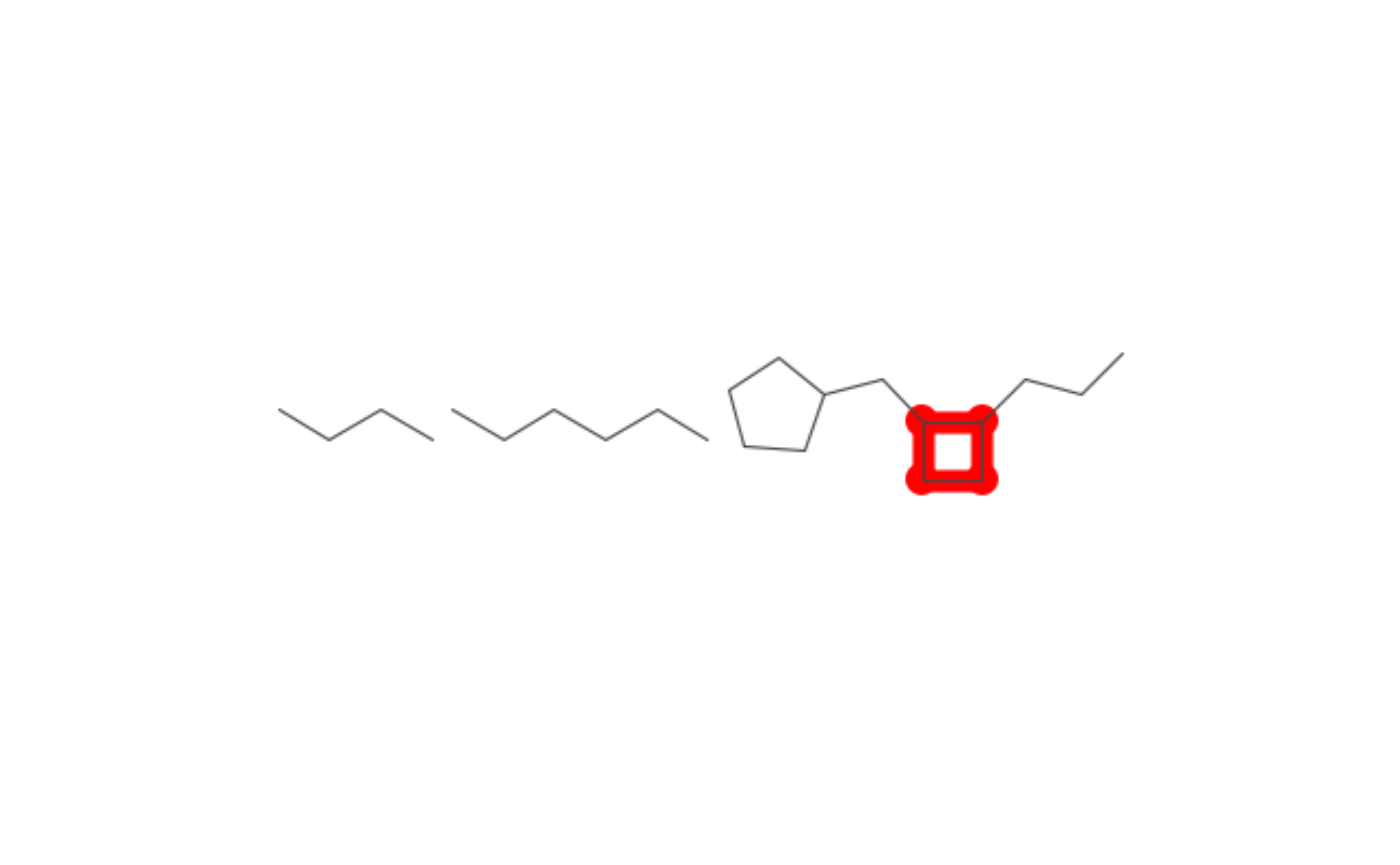
Multiple Colors
mol <- parse_smiles(paste("CC(n1c(C)ncc1c1ccnc(n1)Nc1ccc(cc1)S(=O)(=O)C)C",
"CMGC:CDC2:6gu3:A:FB8", sep=" "))
# imid <- 'c1cnc[nH]1' #1H-imidazole
highlight_imid <- match_smarts("c1cncn1", mol)
highlight_2 <- match_smarts("n1cccnc1Nc2ccccc2", mol)
color <- J("java.awt.Color")
depiction() |>
highlight_atoms(highlight_imid, color$GREEN) |>
highlight_atoms(highlight_2, color$RED) |>
set_size(400, 400) |>
set_zoom(4) |>
outerglow() |>
add_title() |>
depict(mol) |>
get_image() |>
grid::grid.raster()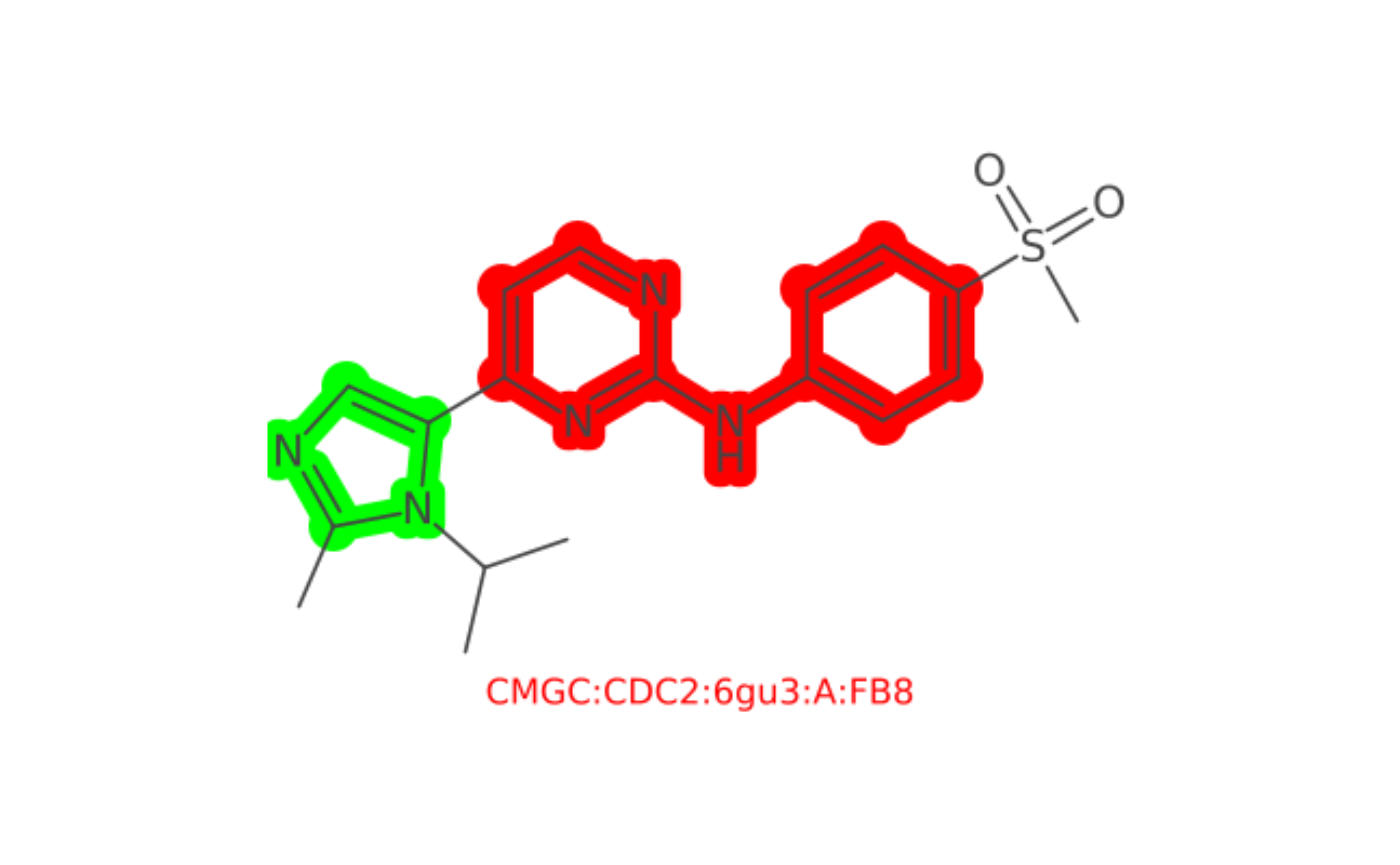
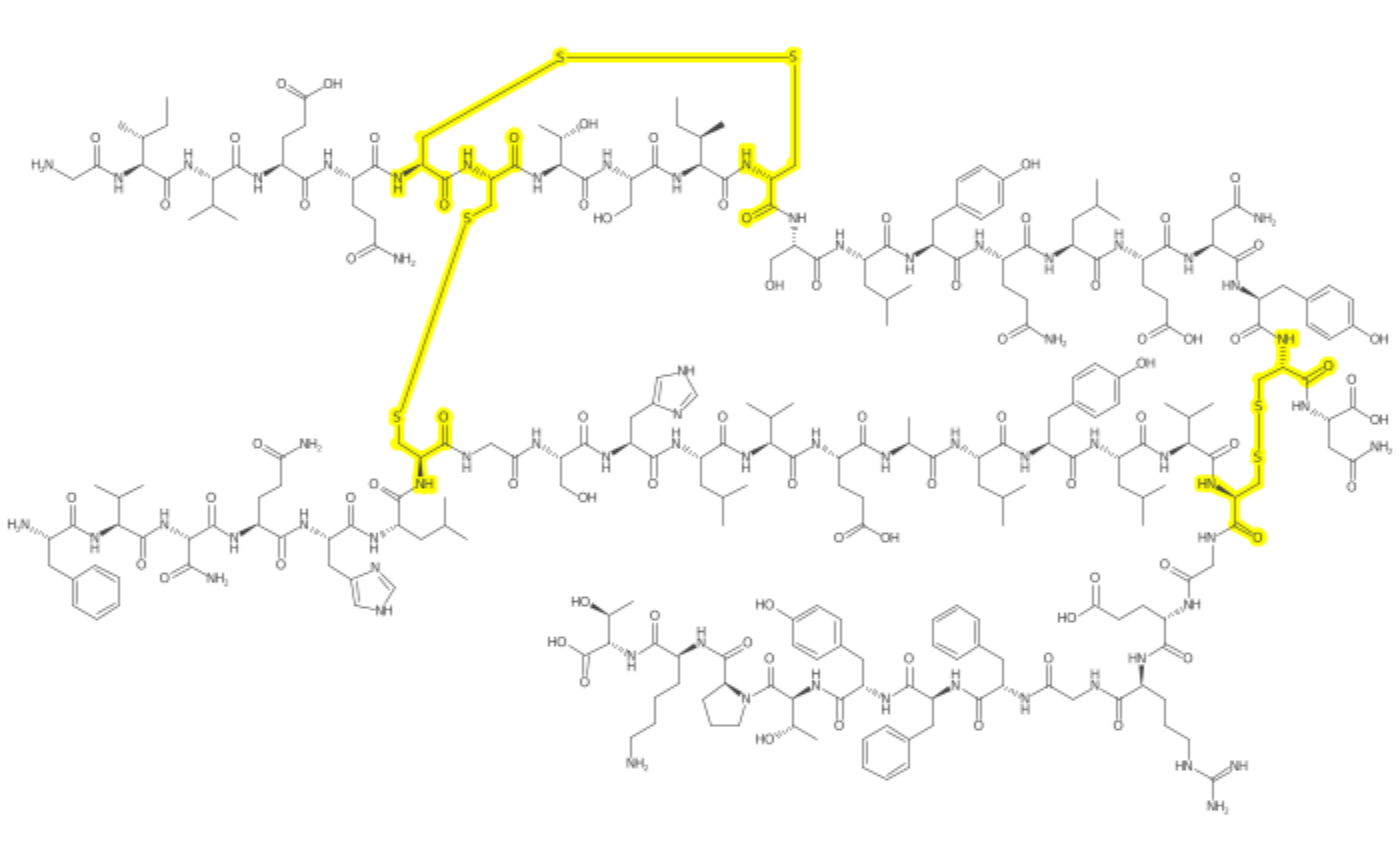
A Larger Example
# laoding a molfile
insulinmol <- system.file("molfiles/ChEBI_5931.mol", package="depict")
insulin <- read_mol(insulinmol)
cysteine <- match_smarts("C(=O)C(CS)N", insulin)
xlinks <- match_smarts("SS", insulin)
depiction()%>%
set_size(700, 400) %>%
set_zoom(10) %>%
outerglow() %>%
highlight_atoms(cysteine, color$YELLOW) %>%
highlight_atoms(xlinks, color$YELLOW) %>%
depict(insulin)%>%
get_image() %>%
grid::grid.raster()